Click on this link for the complete list of publications.
2024
- Harioudh, MK, Perez J, Chong J, Nair S, Lomon So, McCormick KD, Ghosh A, Shao L, Srivastava R, Soveg F, EbertTS, Atianand M, Hornung V, Savan R, Diamond MS, and Sarkar SN. Two distinct mechanisms of virus-specific antiviral activities of Oligoadenylate Synthetase 1 against West Nile virus and SARS-CoV-2. Immunity https://doi.org/10.1016/j.immuni.2024.02.002
- Gokhale NS, Somfleth K, Thompson MG, Sam RK, Marciniak DM, Chu LH, Park M, Dvorkin S, Oberst A, Horner SM,Ong S-E, Gale M Jr., Savan R. Cellular RNA interacts with MAVS to promote antiviral signaling. bioRxiv 2023.09.25.559083; doi: https://doi.org/10.1101/2023.09.25.559083
- Savan R & Gale M Jr. 2023 Innate immunity and interferon in SARS-CoV-2 infection outcome. Immunity 56 (7): 145043-141450. https://doi.org/10.1016/j.immuni.2023.06.018
- So L, Ninomiya K, Hu A, Muir VS, Takamori A, Song J, Buckner JH, Savan R#, and Ziegler SF# (co-corresponding). 2023 Regulatory T cells suppress CD4+ effector T cell activation by controlling protein synthesis. Journal of Experimental Medicine 220 (3): e20221676. https://doi.org/10.1084/jem.20221676. [preprinted 2021 bioRxiv doi.org/10.1101/2021.09.23.461566]
- Kummola L, Salomaa T, Ortutay Z, Savan R, Young HA, Junttila IS. IL-4, IL-13 and IFN-γ -induced genes in highly purified human neutrophils. Cytokine. 2023 Apr;164:156159. https://doi: 10.1016/j.cyto.2023
- Smith JR, Dowling JW, Karp A, Schwerk J, Woodward JJ, Savan R, Forero AR#. 2023. Loss of MEF2A induces replicative stress responses that trigger interferon production. Cell Reports 42(8):112805 doi: 10.1016/j.celrep.2023.112805. [preprinted 2022 bioRxiv https://doi.org/10.1101/2022.09.15.508100 ]
- Tsu BV, Agarwal R, Gokhale NS, Kulsuptrakul J, Ryan AP, Castro LK, Beierschmitt CM, Turcotte E, Fay EJ, Vance RE, Hyde JL, Savan R, Mitchell PS, Daugherty MD. 2023 Host specific sensing of coronaviruses and picornaviruses by the CARD8 inflammasome. PLoS Biology 21(6): e3002144. https://doi.org/10.1371/journal.pbio.3002144 [preprinted 2022 bioRxiv https://doi.org/10.1101/2022.09.21.508960 ]
2022
- Hubbard, N.W., Ames, J.M., Maurano, M. Chu LH, Somfleth KY, Gokhale NS, Werner M, Snyder JM, Lichauco K, Savan R, Stetson DB, Oberst A. ADAR1 mutation causes ZBP1-dependent immunopathology. Nature (2022). https://rdcu.be/cR4z
- Hsieh L, Tu LN, Smith JR, Ghassemian M, Timms A, Savan R, Nigam V. Hemodynamic stress activates inflammatory responses and cell death through spectrin-dependent modulation of Store Operated Calcium Entry. bioRxiv 2022.05.04.490549; doi: https://doi.org/10.1101/2022.05.04.490549
- Hickson SE, Brekke E, Schwerk J, Saluhke I, Zaver S, Woodward J, Savan R, Hyde JL. Sequence diversity in the 3’ untranslated region of alphavirus modulates IFIT2-dependent restriction in a cell type-dependent manner bioRxiv, 11 Dec 2021 DOI: 10.1101/2021.12.10.472177
- Soveg FW, Schwerk J, Gokhale NS, Cerosaletti K, Smith JR, Pairo-Castineira E, Kell AM, Forero A, Zaver SA, Esser-Nobis K, Roby JA, Hsiang T-Y, Ozarkar S, Clingan JM, McAnarney ET, Stone AEL, Malhotra U, Speake C, Perez J, Balu C, Allenspach EJ, Hyde JL, Menachery VD, Sarkar SN, Woodward JJ, Stetson DB, Baillie K, Buckner JH, Gale M Jr, and Savan R. (2021) Endomembrane targeting of human OAS1 p46 augments antiviral activity. Elife. Aug 3;10:e71047. doi: 10.7554/elife.71047 [preprinted bioRxiv, 10.1101/2021.04.21.440697 22 Apr 2021]
- Gokhale NG, Smith JR, Gelder RV, Savan R. 2021 RNA regulatory mechanisms that control antiviral innate immunity. Immunological Reviews. 2021 Aug 17. doi: 10.1111/imr.13019 PMID: 34405416
- Henden AS, Koyama M, Robb RJ, Forero A, Kuns RD, Chang K, Ensbey KS, Varelias A, Kazakoff SH, Waddell N, Clouston AD, Giri R, Begun J, Blazar B R, Degli-Esposti MA, Kotenko SV, Lane SW, Bowerman K, Savan R, Hugenholtz P, Gartlan KH, and Hill GR. (2021) IFNλ Therapy Prevents Severe Gastrointestinal Graft-versus-Host Disease. Blood https://doi.org/10.1182/blood.2020006375
- Allenspach, E. J., Soveg, F., Finn, L. S., So, L., Gorman, J. A., Rosen, A. B. I., Skoda-Smith, S., Wheeler, M. M., Barrow, K. A., Rich, L. M., Debley, J. S., Bamshad, M. J., Nickerson, D. A., Savan, R., Torgerson, T. R., and Rawlings, D. J. (2021) Germline SAMD9L truncation variants trigger global translational repression. Journal of Experimental Medicine 218 (5): e20201195.
- Rommereim LM, Akhade AS, Dutta B, Hutcheon C, Lounsbury NW, Rostomily CC, Savan R, Fraser I, Germain RN, and Subramanian N. (2020) A small sustained increase in NOD1 abundance promotes ligand-independent inflammatory and oncogene transcriptional responses. Science Signaling 13, eaba3244
- Tu LN, Hsieh L, Kajimoto M, Charette K, Kibiryeva N, Forero, A, Hampson SE, Marshall J, O’Brien J, Scatena M, Portman MA, Savan R, Benner C, Aliseda A, Nuri M, Bittel D, Pastuszko P, and Nigam V (2020) Shear stress associated with cardiopulmonary bypass induces expression of inflammatory cytokines and necroptosis in monocytes. JCI Insight https://doi.org/10.1172/jci.insight.141341
- Hendricks MR, Savan R.Interferon-λ at the Center of the Storm. Immunity. 2020 Aug 18;53(2):245-247.
- Schwerk J, Negash A, Savan R, Gale M Jr.Innate Immunity in Hepatitis C Virus Infection. Cold Spring Harb Perspect Med. 2020 Apr 27;a036988.
- Roby JA, Esser-Nobis K, Dewey-Verstelle EC, Fairgrieve MR, Schwerk J, Lu AY, Soveg FW, Hemann EA, Hatfield LD, Keller BC, Shapiro A, Forero A, Stencel-Baerenwald JE, Savan R, Gale M Jr. Flavivirus Nonstructural Protein NS5 Dysregulates HSP90 to Broadly Inhibit JAK/STAT Signaling. Cells 2020 Apr 7;9(4):899.
- Schwerk J, Soveg FW, Ryan AP, Thomas KR, Hatfield LD, Ozarkar S, Forero A, Kell AM, Roby JA, So L, Hyde JL, Gale M Jr, Daugherty MD, Savan R. RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions. Nature Immunology 2019 Dec;20(12):1610-1620.
- Forero A, Ozarkar S, Li H, Lee CH, Hemann EA, Nadjsombati MS, Hendricks MR, So L, Green R, Roy CN, Sarkar SN, von Moltke J, Anderson SK, Gale M Jr, Savan R. Differential Activation of the Transcription Factor IRF1 Underlies the Distinct Immune Responses Elicited by Type I and Type III Interferons. Immunity. 2019 Sep 17;51(3):451-464.e6. doi: 10.1016/j.immuni.2019.07.007. Epub 2019 Aug 27.
- Hemann EA, Green R, Turnbull JB, Langlois RA, Savan R, Gale M Jr. Interferon-λ modulates dendritic cells to facilitate T cell immunity during infection with influenza A virus. Nature Immunology 2019 Aug;20(8):1035-1045. doi: 10.1038/s41590-019-0408-z.
- Soveg F, von Moltke J, Savan R. CIRCling the wagons to protect intestinal stem cells. Nature Immunology 2019 Feb;20(2):114-116. doi: 10.1038/s41590-018-0302-0.
- Joslyn RC, Forero A, Green R, Parker SE, Savan R. Long Noncoding RNA Signatures Induced by Toll-Like Receptor 7 and Type I Interferon Signaling in Activated Human Plasmacytoid Dendritic Cells. J Interferon Cytokine Res. 2018 Sep;38(9):388-405. doi: 10.1089/jir.2018.0086.
- Savan R. Alternative Splicing in Innate Antiviral Immunity. J Interferon Cytokine Res. 2018 Aug;38(8):317-318. doi: 10.1089/jir.2018.29010.rsa.
- Hemann, E.A., Schwerk, J. & Savan, R. (2017). IFN-λ 'guts' neutrophil-mediated inflammation. Nature Immunology 18: 1061–1062. Link
- Forero, A., So, L. & Savan, R. (2017). Re-evaluating strategies to define the immunoregulatory roles of miRNAs.Trends in Immunol. 38(8): 558-566. Link
- Hemann EA, Gale M Jr, Savan R. Interferon Lambda Genetics and Biology in Regulation of Viral Control. Front Immunol. 2017 Dec 6;8:1707. doi: 10.3389/fimmu.2017.01707. eCollection 2017. Review. PubMed PMID: 29270173; PubMed Central PMCID: PMC5723907.
- Jarret, A., McFarland, A.P., Horner, S.M., Kell, A., Schwerk, J., Hong, M., Badil, S., Joslyn, R.C., Baker, D.P., Carrington, M., Hagedorn, C.H., Gale Jr., M. & Savan, R. (2016). Hepatitis-C-virus-induced microRNAs dampen interferon-mediated antiviral signaling. Nature Medicine. 22: 1475–1481. Link Highlighted in the Editor's Choice section of Science Signaling MicroRNAs that interfere with interferon by Alexandra A. Mushegian. Link
- Hong, M.#, Schwerk, J.#, Lim, C.#, Kell, A., Pangallo J, Loo, Y.-M., Jarret, A., Liu, S., Hagedorn, C.H., Gale Jr., M. & Savan, R. (2016). The interferon lambda 4 expression is suppressed by the host during viral infection. (#co-first authors) The Journal of Experimental Medicine Link
- Lim, C., Hong, M. & Savan, R. (2016). The human IL-22 binding protein isoforms are a rheostat for IL-22 signaling.Science Signaling 447:ra95. Link
- Schwerk J. & Savan R. (2015). Translating the Untranslated Region.The Journal of Immunology 195(7): 2963-71. Link
- Schwerk, J.#, Jarret, A.#, Joslyn, R.C. & Savan, R. (2015). Landscape of post-transcriptional gene regulation during hepatitis C virus infection. (#co-first authors) Current Opinion in Virology 12: 75-84. Link
- Lim, C. & Savan, R. (2014). The role of the IL-22/IL-22R1 axis in cancer.Cytokine & Growth Factor Reviews. 25: 257-271. Link
- Savan, R. (2014). Post-Transcriptional Regulation of Interferons and Their Signaling Pathways. Journal of Interferon & Cytokine Research. 34(5): 318-329. Link
- McFarland, A.P., Horner, S.M., Jarret, A., Joslyn, R.C., Bindewald, E., Shapiro, B.A., Delker, D.A., Hagedorn, C.H., Carrington, M., Gale Jr., M. & Savan, R. (2014). The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nature Immunology. 15: 72-79. Link News and Views: Tian, Z. (2014). Outflanking HCV. Nature Immunology. 15: 6-8. Link
- Steinhagen, F.#, McFarland, A.P.#, Rodriguez, L.G., Tewary, P., Jarret, A., Savan, R. & Klinman, D.M. (2013). IRF-5 and NF-kappaB p50 co-regulate IFN-beta and IL-6 expression in TLR9-stimulated human plasmacytoid dendritic cells. (#co-first authors) European Journal of Immunology. 43(7): 1896-1906. Link Commentary: Pelka, K., & Latz, E. (2013). IRF5, IRF8, and IRF7 in human pDCs - the good, the bad, and the insignificant? European Journal of Immunology. 43(7), 1693-1697. Link
- Kulkarni, S.#, Savan, R.#, Qi, Y., Gao, X., Yuki, Y., Bass, S.E., Martin, M.P., Hunt, P., Deeks, S.G., Telenti, A., Pereyra, F., Goldstein, D., Wolinsky, S., Walker, B., Young, H.A. & Carrington, M. (2011). Differential microRNA regulation of HLA-C expression and its association with HIV control. (#co-first authors) Nature. 472(7344): 495-498. Link
- McFarland, A.P., Savan, R., Wagage, S., Addison, A., Ramakrishnan, K., Karwan, M., Duong, T. & Young, H.A. (2011). Localized delivery of interferon-beta by Lactobacillus exacerbates experimental colitis. PLOS One 6(2): e16967. Link
- Savan, R., McFarland, A.P., Reynolds, D.A., Feigenbaum, L., Ramakrishnan, K., Karwan, M., Shirota, H., Klinman, D.M., Dunleavy, K., Pittaluga, S., Anderson, S.K., Donnelly, R.P., Wilson, W.H. & Young, H.A. (2011). A novel role for IL-22R1 as a driver of inflammation. Blood. 117(2): 575-584. Link
Non-inflammatory activities of interferonλ's

Cover art by Falguni Gokhale Foreo et. al. Differential Activation of the Transcription Factor IRF1 Underlies the Distinct Immune Responses Elicited by Type I and Type III Interferons. Immunity 2019 https://doi.org/10.1016/j.immuni.2019.07.007 |
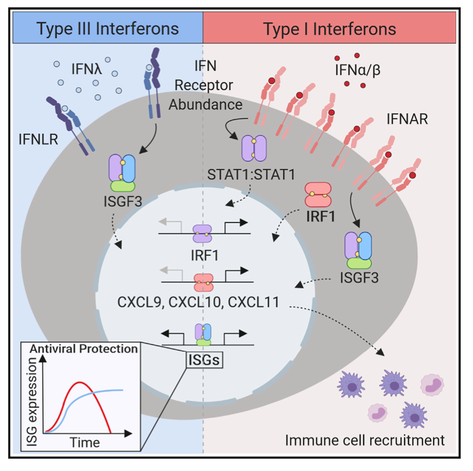
In this study Adriana shows that type I IFNs, but not type III IFNs (IFNλ), promote inflammation at the site of infection. She finds that differential expression of proinflammatory genes results from selective induction of the transcription factor IRF1 by type I IFNs. Type III IFNs induce a tissue repair program, suggesting a division of labor that spans proinflammatory and tissue repair functions to promote protective immunity. A commentary by Casazza and Lazear discusses the implications of our work. |
Variation in the interferon genes determine susceptibility to Hepatitis C virus infection
|
Genome-wide association studies identified a strong association between polymorphisms near IFNL3 and clearance of HCV. In the first study, we reported the identification of a functional polymorphism (rs4803217) in the 3' untranslated region (UTR) of IFNL3 mRNA that dictated transcript stability. We found that this polymorphism influenced AU-rich element-mediated decay of IFNL3 mRNA, as well as the binding of HCV-induced microRNAs during infection. We show that HCV induces aberrant expression of two host microRNAs, miR-208b and miR-499a-5p, encoded by myosin genes in infected hepatocytes. These miRNAs, along with AMD, suppress IFNL2 and IFNL3 gene expression, to support viral persistence. Together, these destabilizing mechanisms mediated robust mRNA repression of the unfavorable IFNL3 polymorphism. This study was highlighted by Tian, Asselah, and Frankfurter Aligemeine Zeitung.
In the second study, we show myomiRs also dampen type I IFN signaling in HCV-infected hepatocytes by directly down-regulating expression of the type I IFN receptor chain, IFNAR1. Inhibition of these miRNAs by using miRNA inhibitors during HCV infection increased expression of IFNAR1. Additionally, inhibition rescued the antiviral response to exogenous type I IFN, as measured by a marked increase in IFN stimulated genes and a decrease in HCV load. Treatment of HCV-infected hepatocytes with type I IFN increased expression of myosins over HCV infection alone. Since these miRNAs can suppress IFNl2 and IFNL3, these data collectively define a novel cross-regulation between type I and III IFNs during HCV infection. Thus, we propose myomiR regulation of IFNL3 as a mechanism to explain why interferon lambda genotype associates with response to type I IFN-based therapies in HCV-infected patients. This study was highlighted at Science Signaling by Alexandra Mushegian. |
An antiviral protein that protects against severe COVID-19 disease
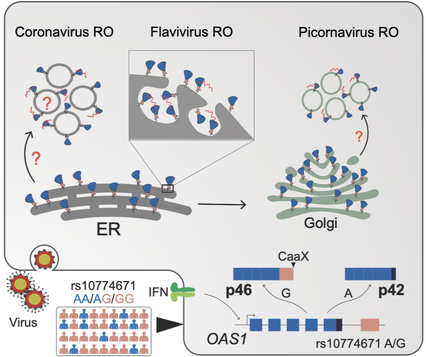
In this study, Frank and Hannes show that the prenylated isoform of oligoadenylate synthetase 1 (OAS1 p46) neutralizes positive-strand RNA viruses that cause severe human diseases.
Prenylated OAS1 accesses the viral replication sites and activates an enzyme called RNaseL that chews up the genetic material of flaviviruses, picornaviruses,
and SARS-CoV-2. We also show that a splice-site variation in human OAS1 is responsible for the production of the prenylated OAS1 isoform, and this correlates with protection from severe COVID-19 disease.
An article in the Atlantic titled "A Better Way to Think About Your Risk for COVID-19" by Roxanne Khamsi highlights our study. This article also highlights how innate immune genes play a critical role in early immune responses to control SARS-CoV-2.
The University of Washington, Science in Seattle and a local new channel discuss our findings.
Prenylated OAS1 accesses the viral replication sites and activates an enzyme called RNaseL that chews up the genetic material of flaviviruses, picornaviruses,
and SARS-CoV-2. We also show that a splice-site variation in human OAS1 is responsible for the production of the prenylated OAS1 isoform, and this correlates with protection from severe COVID-19 disease.
An article in the Atlantic titled "A Better Way to Think About Your Risk for COVID-19" by Roxanne Khamsi highlights our study. This article also highlights how innate immune genes play a critical role in early immune responses to control SARS-CoV-2.
The University of Washington, Science in Seattle and a local new channel discuss our findings.
Isoform-specific functions of zinc-finger antiviral protein
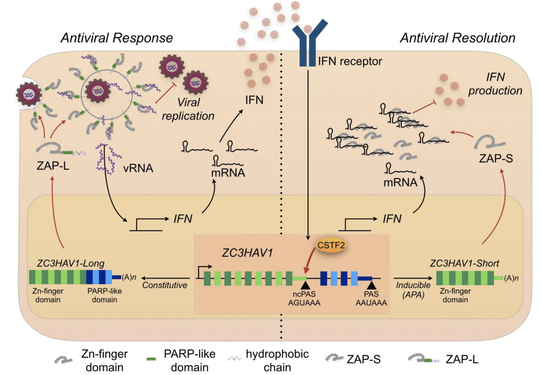
A study from Hannes and Frank shows that the isoforms of the RNA-binding protein ZAP have distinct functions. The long-isoform functions as a virus restriction factor, and the short isoform is an interferon-resolution factor. We found that the specific functions are tied to their subcellular localization.
An editorial by Dr. Erin Williams. at Science Signaling and Science in Seattle highlighted the novel findings of this work.




